Research Field
Establishment of early warning system for influenza epidemic
Article source: Date time:2009-11-09
Study purposes
The prediction of influenza epidemic is the key to prevent and control influenza. Many factors affect the prediction including transmissibility, viral pathogenicity, susceptibility and host immunity along with other natural factors. Current prediction system often relies on experience and conjecture due to technological limitations. Establishment of an effective early warning system for influenza epidemic is still a global challenge. The purpose of this study is to establish an early warning model and develop analytic software, based on the established data and new modeling method of biology and epidemiology, for rapid detection of antigenicity, transmissibility and influenza viral pathogenicity; to further optimize and evaluate the model and data according to the requirements of modeling; and eventually apply this system in practice. This study is a collaborative project of
Study schemes

The technology roadmap of early warning system
Study progress
The HA1 protein sequences of all human influenza H3N2 were downloaded from GenBank. Based on the influenza surveillance network of
Based on experience and sequence analysis, 12 characteristics that reflect the changes of antigen relationship were acquired and discretized into four groups. Bayesian antigen predictive model and antigen network were established for antigen cluster analysis. The resulted figures were used to reflect virus vairation and describe the transmission pattern of influenza virus. The established model and network were eventually applied in recommendatioon of candidate strains for vaccine production. Fig.3-63 B shows the result of computer simulation in comparison with that of WHO recommended method.
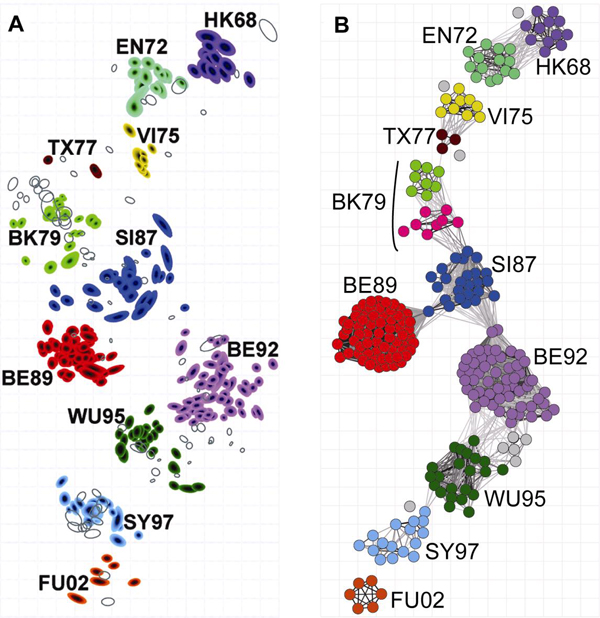
Candidate strains for vaccine production selected by WHO recommended HI test ( A) and computer simulation (B).
A. WHO recommended candidate vaccine strains by HI test;
B. Computer simulation antigenic variation network by sequence
Conclusions
Predictive computer model of virus antigenic variation has been established based on the HA1 sequence. The preliminary results indicate that this model can be used to select influenza candidate vaccine strains, and the predicted viral sequence is in agreement with that selected by WHO recommended HI test.
Future study
i. To further validate and optimize all parameters used in the model in the laboratory;
ii. To develop relevant software and early warning platform for automatic screening of candidate vaccine strain and early warning of influenza epidemic.